Projects
How it all began… Tumor budding in colorectal cancer
ngTMA® was driven by our motivation to study protein expression in tumor buds in colorectal cancers. Tumor buds are single cells or small tumor cell clusters found either at the invasion front or within the main tumor body of many different cancer types. Convinced that these cells were a cause of distant metastatic spread, we wanted to characterize tumor buds on a protein and molecular level and relate the differences in expression back to patient outcome. There was no way to do this using standard tissue microarraying techniques. In 2011, we bought a slide scanner and our first tissue mciroarraying device. Finally, we could annotate a digital slide and align this scan with the image of the corresponding block to be able to core out areas of budding! The rest, as they say, is history… We trademarked our ngTMA® brand in 2012 and since then have helped achieve the vision of more than 150 projects.
ngTMA® today and future…. Digital Pathology - Coming soon!
#deeplearning; #AI, #computational, #machinelearning, #digital
Our research group applies an interdisciplinary approach to colorectal cancer. We investigate the relevance of tumor budding and the tumor microenvironment in different clinical scenarios, and apply computational methods (e.g. deep learning, graphs) to discover new features, train models and eventually validate and translate findings in routine setting.
Other projects ngTMA based
Prognostic and Biomarkers Studies
Rodriguez A et al, BioRxiv (2020) doi: 10.1101/2020.05.12.092296v1
Multi-regional, genomic, and targeted transcriptomic analysis using ngTMA® technology
Using ngTMAs and tissue punches, this study demonstrated that prostate cancer brain metastases have genomic dependencies that may be exploitable through clinical interventions
Zeng et al, Nature (2019) doi: 10.1038/s41586-019-1576-6
ngTMA® in journal Nature
Our technology was necessary to compare the protein expression (GluN2B-mediated NMDAR signalling) in paired breast primary tumors and brain metastasis.
Litle NK et al, Cell (2019) doi: 10.1016/j.cell.2019.03.010
ngTMA® in journal Cell
RORγ expression in PanIN lesions and nuclear RORγ localization in invasive carcinoma could be useful markers to predict PDAC aggressiveness.
Langer R et al, Cancers (2018) doi: 10.3390/cancers10090281
Autophagy markers can be analysed using ngTMA®
Different states of autophagy characterized by distinct p62 and LC3B expression patterns may be linked to patient's prognosis in pSQCC.
Schurch et al. OncoImmunology (2018) doi: 10.1080/2162402X.2017.1373235
ngTMA® collected 45 mesotheliomas to evaluate the CD47 signal
This study demonstrated that CD47 is an accurate novel diagnostic DMM biomarker and that blocking CD47 may represent a promising therapeutic strategy for DMM.
Galvan et al, Virchows Archiv (2019) doi: 10.1007/s00428-019-02714-6
Analyzing cancer-associated fibroblasts…
We were able to collect samples of primary resected tumors treated with neoadjuvant therapy from esophageal adenocarcinoma patients to investigate the role of cancer-associated fibroblasts (CAFs) in response to therapy.
Wyss et al , Clin Colorectal Cancer (2019) doi: 10.1016/j.clcc.2018.09.007
PD-L1 in CRC can be studied using ngTMA®
The main objective for the future is to identify patients who will benefit most from immunotherapy
Morteza Seyed Jafary S et al PlosOne (2018) doi: 10.1371/journal.pone.0207019
ngTMA® collected 238 samples of melanocytic nevi, primary and metastatic melanomas
The study showed that VEGF-C and VEGF-R2 might represent new prognostic marker in Melanoma.
Establishment of New Techniques
Image courtesy of Fabienne Birrer and Tess Brody, UniBern (Hyperion)
Adapting layouts for protocols establishment on Imaging mass cytometry
Schurch CM et al, BioRxiv (2019) doi: https://doi.org/10.1101/743989, 2019
Adapting layouts to new CODEX technology
Our layouts and TMAs can be designed for 13x10mm flow cell for CODEX Technology. That means 100 punches per TMA block.
Our biggest layout… 420 punches per TMA block!
Project performed with 3’780 FFPE donor blocks in 2018. In total, we were able to construct 72 TMA blocks with 29’782 punches from patients diagnosed with colorectal cancer.
Original idea from: Frosina & Jungbruth, Appl Immunohistochem Mol Morphol (2016) doi: 10.1097/PAI.0000000000000239
Developing new controls for IHC
By using some tissues as carriers (i.e. liver), as negative controls and inserting other tissues as positive controls, we can optimize our immunostainings.
Vasella et al. Microarrays (2015) doi.org/10.3390/microarrays4020188
ngTMA® for Molecular applications
Our instrumentation is appropriate for use as an accessory to molecular applications. No cross-contamination appears to occur between samples punched with the same device, although a cleaning step in between donor blocks is still recommended.
Watenberg M et al, Eur J Cancer (2016) doi: 10.1016/j.ejca.2016.06.013
ngTMA® was used for ISH and FISH in pancreatic ductal adenocarcinoma
PTEN deletion is a major cause of PTEN protein loss in PDAC and correlates with aggressive characteristics and worse outcome.
Meyer et al, Human Patholgoy (2019) doi: 10.1016/j.humpath.2019.02.002
A ngTMA® of matched Pan-CK-positive and -negative stroma
CK+/VIM+ tumor cells could represent a subgroup of tumor buds in partial EMT. CK+/VIM+ stroma may be of mesothelial origin and shows features of mesenchymal cells and cancer-associated fibroblasts.
Digital Pathology Studies
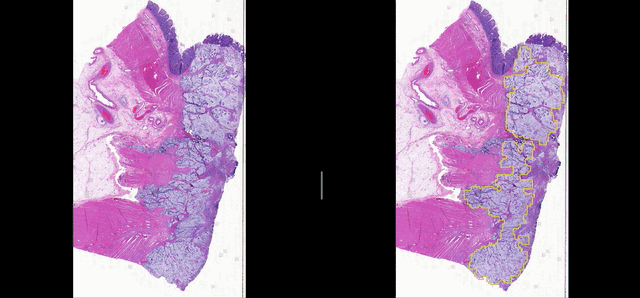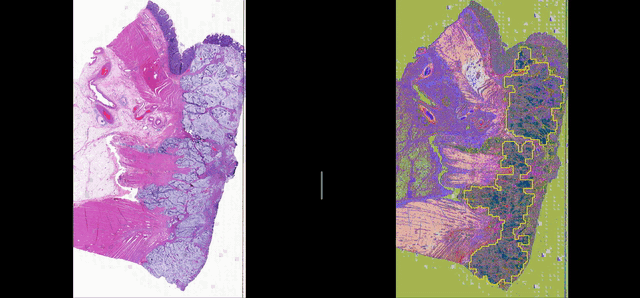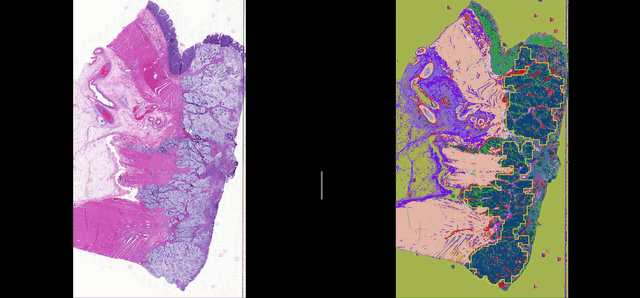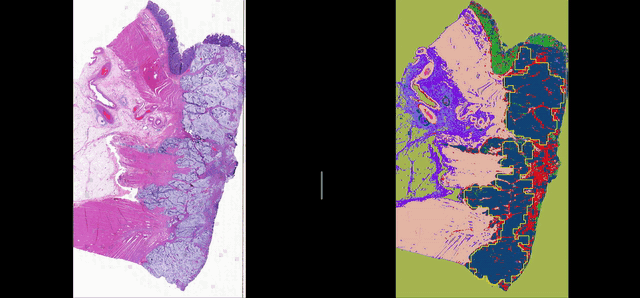
Deep learning for Group Affinity Weakly Supervised Segmentation
A precise tissue segmentation of histopathology images using weakly supervised deep learning was proposed to support and improve the pathologist’s workflow by eliminating inter- and intra-observer variability. It was validated with a colorectal cancer dataset with 163 H&E WSIs from 97 patients.
Computational Analysis of Colorectal Cancer Metastasis in Lymph Nodes
How QuPath works on ngTMAs...
QuPath is an open, powerful, flexible, extensible software platform for whole slide image and TMA analysis.
Nguyen et al, Sci Rep. (2021) doi: 10.1038/s41598-021-81352-y
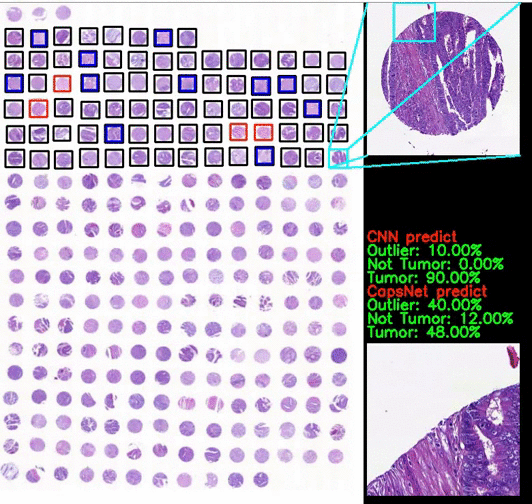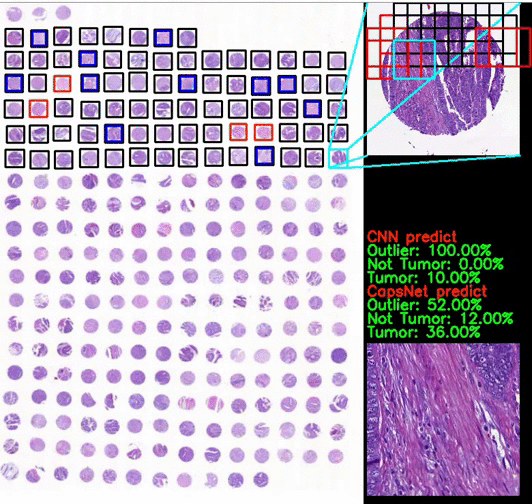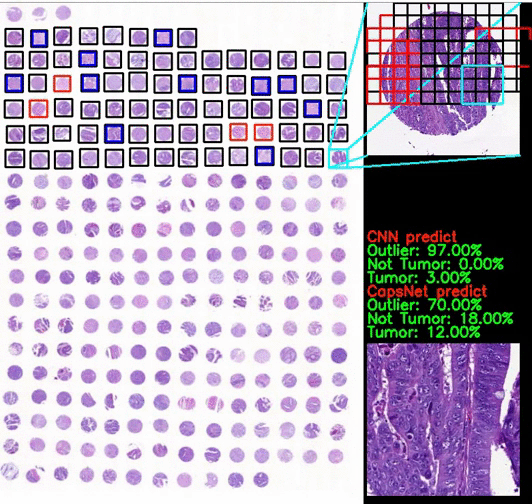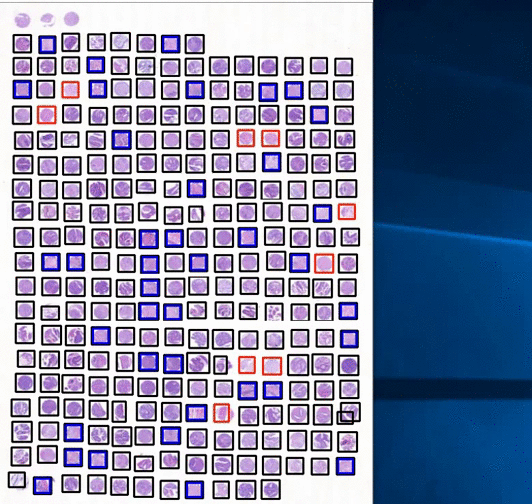
Classification of CRC images from ngTMAs by ensemble deep learning methods
Using high-accuracy algorithm for colorectal tissue classification in high-throughput ngTMAs is amenable to images from different institutions, core sizes and stain intensity. It helps to reduce error in ngTMA core evaluations with previously given labels.
Screening of metabolomics markers expression using ngTMA®
Zahnd S et al. J Pathol Inform (2019) doi: 10.4103/jpi.jpi_65_18
Digital image analysis (DIA) workflow as part of ngTMA®
Cathepsin B expression analyzed by QuPath within the tumor epithelial compartment was identified as a strong feature of less aggressive tumor behavior and favorable outcome.
Nolte S et al. J of Pathology (2016) doi: 10.1002/cjp2.62
Combining ngTMA® and image analysis in a colon cancer cohort of 612 patients
The aim was to investigate CDX2 and its counterpart CDX1 using a combined ngTMA® and digital image analysis approach to evaluate issues of accuracy of coring, intra‐tumoural heterogeneity and prognosis in a large cohort of 612 colorectal cancer patients
Scorenado: an efficient and user-friendly visual assessment tool.
Incorporating Scorenado into the research routine not only allows researchers to efficiently evaluate TMA and whole slides but also greatly facilitates the creation of expert annotated datasets on the fly, which may have the potential to significantly advance digital clinical histopathology.